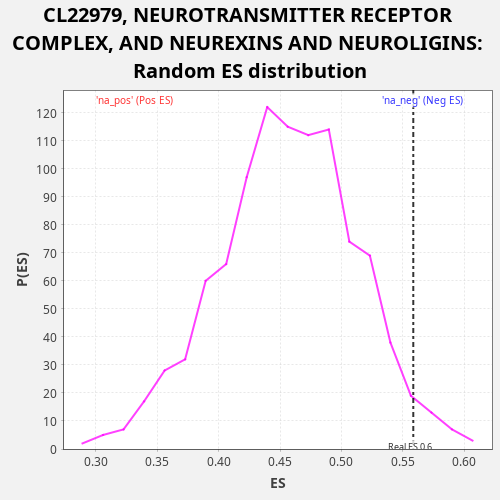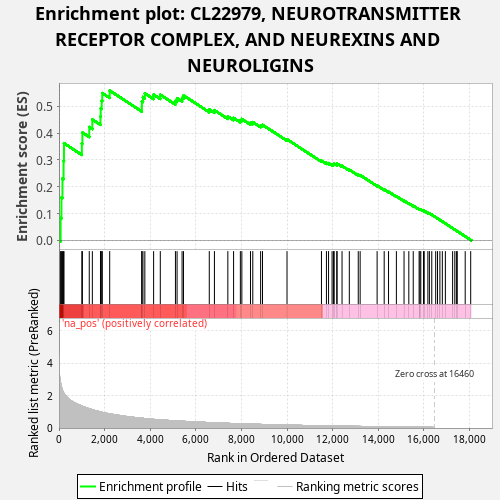
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Medication for pain relief, constipation, heartburn__6154-both_sexes-4 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.5585215 |
| Normalized Enrichment Score (NES) | 1.2240598 |
| Nominal p-value | 0.026 |
| FDR q-value | 0.41262087 |
| FWER p-Value | 0.964 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CHRM3 | 70 | 2.772 | 0.0834 | Yes |
| 2 | DLGAP4 | 107 | 2.513 | 0.1605 | Yes |
| 3 | GRM8 | 152 | 2.310 | 0.2307 | Yes |
| 4 | GRIK1 | 200 | 2.164 | 0.2963 | Yes |
| 5 | LRRTM1 | 218 | 2.126 | 0.3622 | Yes |
| 6 | OLFM2 | 1002 | 1.348 | 0.3612 | Yes |
| 7 | DLGAP3 | 1026 | 1.332 | 0.4018 | Yes |
| 8 | GRIN2A | 1327 | 1.185 | 0.4225 | Yes |
| 9 | GRIK4 | 1462 | 1.126 | 0.4505 | Yes |
| 10 | GRM1 | 1819 | 0.986 | 0.4618 | Yes |
| 11 | GRIN3B | 1828 | 0.983 | 0.4923 | Yes |
| 12 | NLGN1 | 1872 | 0.968 | 0.5204 | Yes |
| 13 | CBLN2 | 1898 | 0.959 | 0.5491 | Yes |
| 14 | GRIK2 | 2223 | 0.869 | 0.5585 | Yes |
| 15 | FBXO40 | 3632 | 0.603 | 0.4993 | No |
| 16 | CACNG8 | 3633 | 0.603 | 0.5183 | No |
| 17 | HOMER2 | 3681 | 0.596 | 0.5344 | No |
| 18 | SLITRK5 | 3764 | 0.582 | 0.5482 | No |
| 19 | GRIN2D | 4149 | 0.530 | 0.5436 | No |
| 20 | NLGN2 | 4442 | 0.500 | 0.5431 | No |
| 21 | HTR3E | 5106 | 0.441 | 0.5201 | No |
| 22 | HTR3D | 5178 | 0.434 | 0.5299 | No |
| 23 | CNTN4 | 5397 | 0.415 | 0.5308 | No |
| 24 | NETO2 | 5460 | 0.410 | 0.5403 | No |
| 25 | HTR3B | 6590 | 0.332 | 0.4880 | No |
| 26 | CHRM5 | 6816 | 0.316 | 0.4855 | No |
| 27 | CLSTN3 | 7404 | 0.285 | 0.4619 | No |
| 28 | CACNG5 | 7656 | 0.271 | 0.4565 | No |
| 29 | DLG2 | 7953 | 0.256 | 0.4481 | No |
| 30 | HOMER3 | 8018 | 0.252 | 0.4525 | No |
| 31 | SHISA6 | 8397 | 0.237 | 0.4390 | No |
| 32 | NETO1 | 8498 | 0.232 | 0.4407 | No |
| 33 | CDH9 | 8842 | 0.219 | 0.4286 | No |
| 34 | GRIA1 | 8924 | 0.216 | 0.4309 | No |
| 35 | CNIH3 | 9999 | 0.177 | 0.3768 | No |
| 36 | SYNGAP1 | 11509 | 0.133 | 0.2972 | No |
| 37 | HTR3C | 11728 | 0.128 | 0.2891 | No |
| 38 | NRXN2 | 11821 | 0.126 | 0.2880 | No |
| 39 | SHANK1 | 11975 | 0.122 | 0.2833 | No |
| 40 | DLG4 | 12035 | 0.121 | 0.2839 | No |
| 41 | GRIK3 | 12067 | 0.120 | 0.2859 | No |
| 42 | GRIA4 | 12183 | 0.118 | 0.2833 | No |
| 43 | GRM5 | 12193 | 0.117 | 0.2865 | No |
| 44 | GRIN3A | 12412 | 0.112 | 0.2779 | No |
| 45 | GRID1 | 12731 | 0.105 | 0.2636 | No |
| 46 | SHISA7 | 13123 | 0.096 | 0.2449 | No |
| 47 | TJAP1 | 13206 | 0.095 | 0.2433 | No |
| 48 | NRXN3 | 13950 | 0.081 | 0.2046 | No |
| 49 | SHANK3 | 14260 | 0.075 | 0.1898 | No |
| 50 | GRIA2 | 14451 | 0.072 | 0.1815 | No |
| 51 | HOMER1 | 14795 | 0.066 | 0.1646 | No |
| 52 | GRM7 | 15131 | 0.061 | 0.1479 | No |
| 53 | SHANK2 | 15340 | 0.058 | 0.1382 | No |
| 54 | VWC2L | 15534 | 0.054 | 0.1291 | No |
| 55 | NRXN1 | 15792 | 0.050 | 0.1164 | No |
| 56 | SHISA9 | 15828 | 0.049 | 0.1160 | No |
| 57 | LHFPL4 | 15866 | 0.049 | 0.1155 | No |
| 58 | DLGAP1 | 15984 | 0.046 | 0.1105 | No |
| 59 | HTR3A | 16015 | 0.046 | 0.1103 | No |
| 60 | CNIH2 | 16176 | 0.043 | 0.1027 | No |
| 61 | NLGN4X | 16243 | 0.041 | 0.1004 | No |
| 62 | GRIN1 | 16347 | 0.037 | 0.0958 | No |
| 63 | CBLN4 | 16519 | -0.000 | 0.0863 | No |
| 64 | DLG3 | 16599 | -0.000 | 0.0819 | No |
| 65 | CBLN1 | 16703 | -0.000 | 0.0762 | No |
| 66 | MDGA1 | 16810 | -0.000 | 0.0703 | No |
| 67 | GRM4 | 16943 | -0.000 | 0.0630 | No |
| 68 | GRID2 | 17261 | -0.000 | 0.0454 | No |
| 69 | GRIN2C | 17357 | -0.000 | 0.0401 | No |
| 70 | PTCHD1 | 17427 | -0.000 | 0.0363 | No |
| 71 | CACNG2 | 17465 | -0.000 | 0.0343 | No |
| 72 | NLGN3 | 17813 | -0.000 | 0.0150 | No |
| 73 | GRIN2B | 18054 | -0.000 | 0.0017 | No |